Structured illumination microscopy (SIM) has emerged as the most powerful super-resolution technique for live cell imaging, owing to its inherent advantages of high speed, low phototoxicity, and compatibility with various dyes. With the advancement of SIM, researchers have proposed a range of algorithms such as Open-SIM, fairSIM, Hessian-SIM, HiFi-SIM, etc. These open-source software solutions have also spurred hardware innovations, including SLM-SIM, DMD-SIM and commercial systems such as Airy-SIM. The convergence of algorithms and hardware has fostered an open and collaborative environment in the field of SIM.
In 2008, Gustafsson introduced the concept of 3D structured illumination microscopy (3DSIM), which offers twice the axial resolution of 2D SIM, effectively eliminating common defocus artifacts encountered in 2D SIM and enabling whole-cell imaging. However, due to its inherent complexity, the development of 3DSIM has lagged behind its 2D counterpart. Existing 3DSIM algorithms are either proprietary in commercial systems or rely on traditional Weiner-3DSIM approaches, resulting in limited user-friendliness and the presence of significant artifacts. Consequently, there is an urgent need for an open-source and robust 3DSIM software to meet the growing demands for further advancements in this field.
Building upon their previously proposed 2DSIM reconstruction platform, Open-SIM (IEEE JSTQE 2018), Peng Xi's research group has recently introduced a new open-source 3DSIM reconstruction platform, Open-3DSIM, which has been published in the prestigious journal Nature Methods. In contrast to Open-SIM, Open-3DSIM provides three platforms—MATLAB, Fiji, and Exe—to cater to diverse user requirements. It offers high-fidelity reconstruction with minimal artifacts, even under low signal-to-noise ratio (SNR) conditions. Notably, Open-3DSIM incorporates dipole orientation imaging, expanding its capabilities to include polarization imaging.
To enhance the performance of Open-3DSIM, Peng Xi's research group has devised an adaptive parameter estimation method that collaboratively estimates parameters using ±first and ±second frequency peaks. This novel approach significantly improves the accuracy of 3DSIM parameter estimation, particularly under challenging conditions of low SNR and low modulation. Moreover, a series of frequency domain filters have been designed to suppress artifacts and preserve weak information. These include notch functions to mitigate hexagonal artifacts, primary filtering functions to suppress sidelobes and ringing artifacts, and secondary filtering functions to retain delicate details. These optimizations ensure faithful reconstructions while effectively suppressing defocus background and various artifacts. Consequently, Open-3DSIM exhibits comprehensive superiority over existing reconstruction algorithms (AO-3DSIM, 4BSIM, SIMnoise, etc.) and commercial reconstruction software (e.g., GE | OMX). Notably, under extremely low SNR conditions, Open-3DSIM demonstrates similar mean squared error (MAE) and signal-to-noise ratio (SNR) to other algorithms, highlighting its exceptional performance in low SNR scenarios.
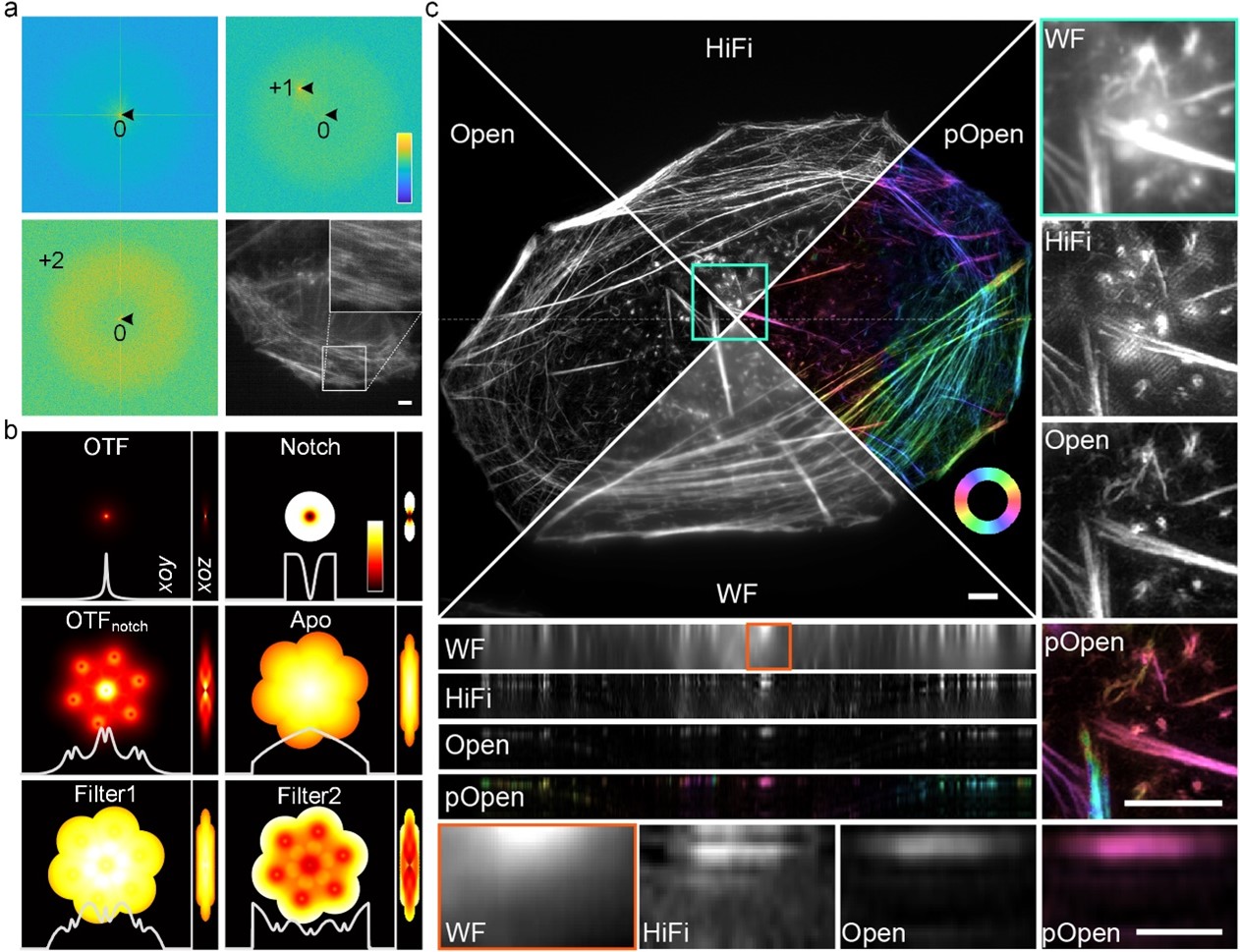
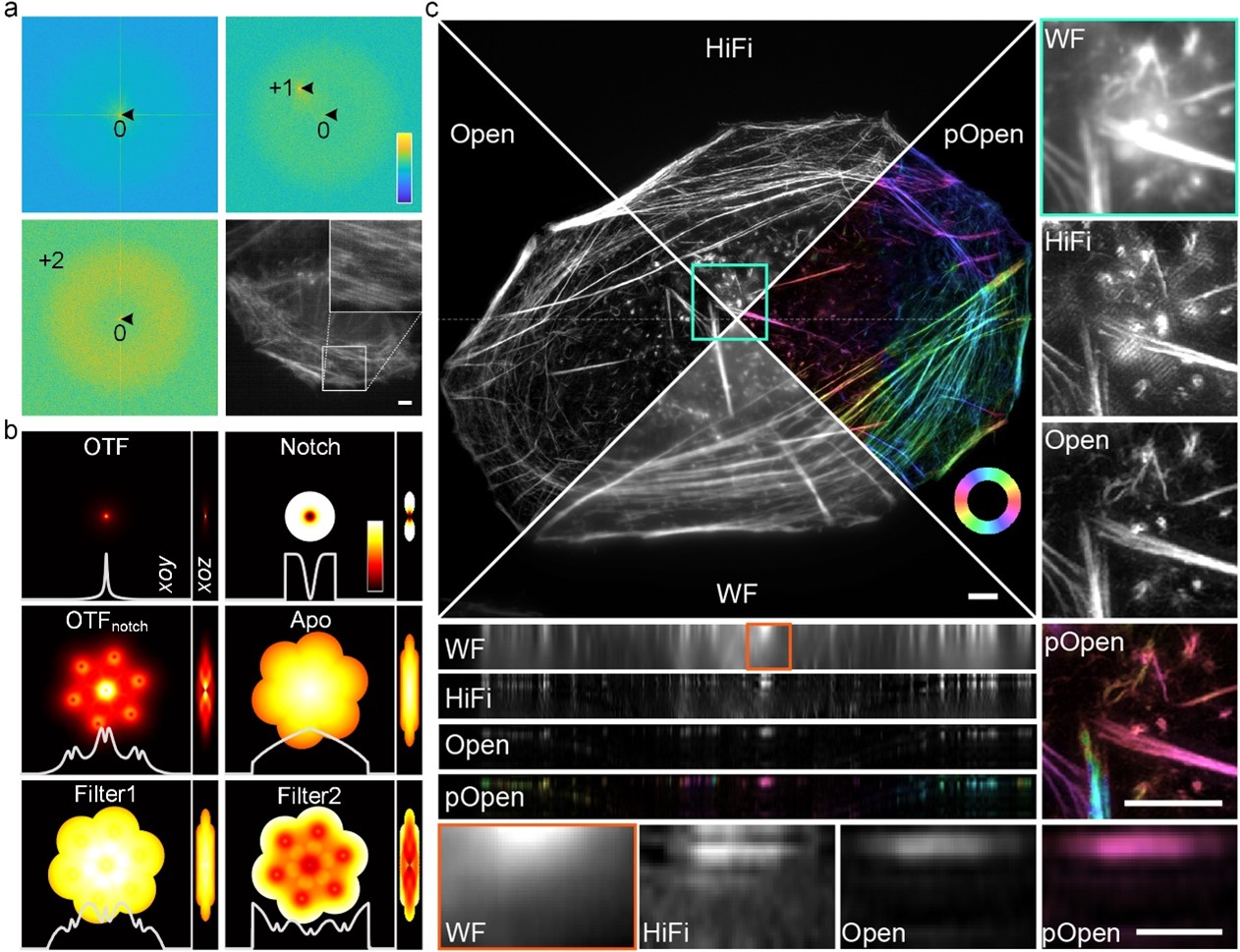
Figure 1. a, Adaptive parameter estimation using ±first and ±second frequency peaks under low signal-to-noise ratio and low-modulation conditions. b. Various spectrum optimization filters designed based on the optical transfer function and estimated parameters. c. Comparison of reconstruction on actin filament using wide field imaging (WF), single layer 3DSIM algorithm (such as HiFi-SIM), Open-3DSIM, and dipole orientation imaging of Open-3DSIM (pOpen).
Furthermore, Peng Xi's research group has introduced polarization information into the post-processing stage of 3DSIM reconstruction, enabling the acquisition of dipole orientation maps for various biological samples without requiring any hardware modifications. As illustrated in Figure 2, the exceptional qualities of Open-3DSIM, including its low artifact levels and high resolution, have facilitated successful three-color imaging reconstructions of Cos-7 cells. Moreover, the research team analyzed the three-dimensional structure of mitochondria, capturing their dynamic processes of separation and fusion. Additionally, in 5 μm thick mouse kidney slices, they obtained valuable three-dimensional dipole orientation information of microfilament organelles. Open-3DSIM encompasses six dimensions (XYZλθT), enabling super-resolution imaging in both lateral and vertical directions, multi-color imaging, time-lapse observations, and dipole orientation analysis. This comprehensive capability establishes Open-3DSIM as a valuable tool for multidimensional imaging studies of subcellular organelles.
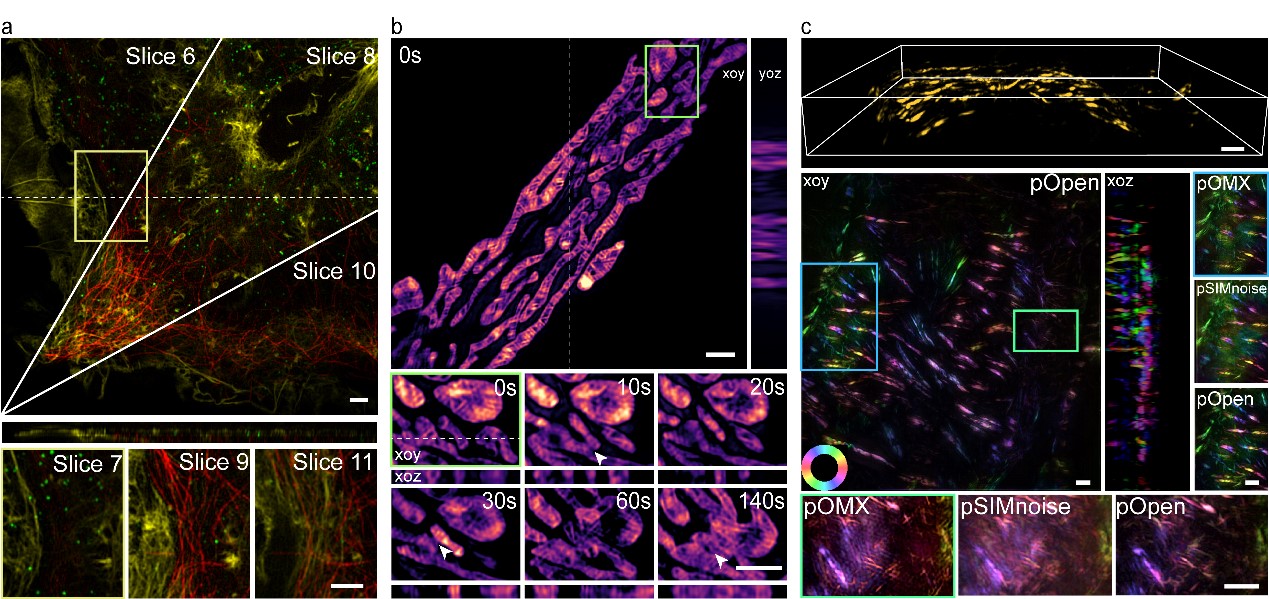
Figure 2. Open-3DSIM establishes a six-dimensional imaging mode with lateral/vertical super-resolution, multi-color, time-lapse, and dipole orientation. a. Reconstruct for multi-color Cos-7 cells, b, Analysis of the three-dimensional mitochondrial division, fusion, apoptosis, and c, Obtain polarization information on the actin filament of mouse kidney slices.
Open-3DSIM exhibits a remarkable versatility by being compatible not only with a single microscopy platform but also with a wide range of commercial or custom-built 3DSIM systems, including but not limited to GE|OMX and Nikon|N-SIM. Furthermore, it seamlessly integrates with diverse image optimization techniques based on regularization, deconvolution, or machine learning. This compatibility, combined with its exceptional performance, modular design, and exceptional user-friendliness, establishes Open-3DSIM as a robust software foundation for the next generation of 3DSIM development.
Peng Xi envisions Open-3DSIM to serve as the international standard for 3DSIM reconstruction, thereby catalyzing the further advancement of multi-dimensional live cell super-resolution imaging technology. Through its widespread adoption, Peng Xi aims to foster collaborative research and promote the rapid progress of this transformative field.
This work was published online in Nature Methods on July 20, 2023, with Ruijie Cao, a PhD candidate from the School of Future Technology at Peking University, as the first author. Prof. Peng Xi is the corresponding author of the paper. The authors thank National Center for Protein Sciences at Peking University, and Dr. Christopher Leterrier from Aix Marseille University for providing the N-SIM data for the paper. The author are also grateful to Professor Li Hui (Suzhou Institute of Biomedical Engineering, Chinese Academy of Sciences), Professor Marcel Mueller (KU Leuven), and Professor Lin Shao (Yale University) or their invaluable expertise, insightful review, and constructive comments that greatly contributed to the refinement of this article.
This work has been supported by projects of the National Natural Science Foundation of China and National Key R&D Program of China.