In mammals, hematopoietic stem cells have the ability of self-renewal and can differentiate into multiple cell types in blood, maintaining cell populations and functions of the hematopoietic system. Although definitive long-term hematopoietic stem cells (LT-HSCs) are known to emerge via endothelial-to-hematopoietic transition (EHT) through lineage tracing and single-cell transcriptome sequencing1, 2, it remains elusive that how the multi-layered epigenome is sequentially unfolded in a small portion of endothelial cells (ECs) transitioning into the hematopoietic fate.
On Jan. 17th, 2022, Aibin He Lab from College of Future Technology in Peking University and Peking University-Tsinghua University Joint Center for Life Sciences, published the work “ Pre-configuring chromatin architecture with histone modifications guides hematopoietic stem cell formation in mouse embryos ” online in Nature Communications .
The scientific question in this study is how multimodal epigenetic mechanisms promotes LT-HSCs differentiating from early arterial ECs (eAECs) in mammals.
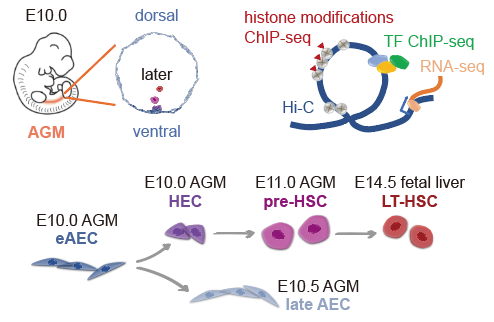
With optimized cell-surface markers, eAECs, hemogenic endothelial cells (HECs), pre-hematopoietic stem cells (pre-HSCs) and LT-HSCs were collected from aorta-gonad-mesonephros and livers from mouse embryos. Low-input itChIP-seq (indexing and tagmentation ChIP-seq, by Aibin He Lab before)3 and sisHi-C (small-scale in situ Hi-C)4 were performed to identify chromatin structures, histone modifications and hematopoiesis-related transcription factor (TF) RUNX1 occupancy in the whole genome (Figure 1), followed by integrated analysis of published single-cell RNA-seq data on the same four cell populations1, 2.
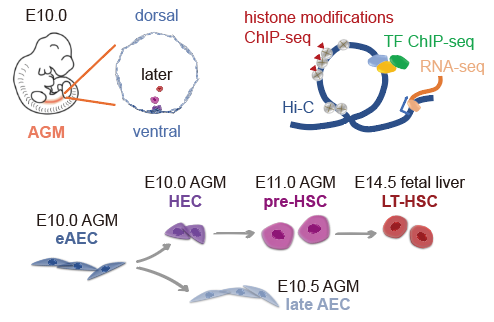
Figure 1. Schematic of experimental design
There is an enhanced intra-TAD (topologically associating domain) connectivity associated with hematopoiesis. At a large-scale view, only 10.78% of genomic regions displayed A/B compartment flipping across the four populations. Those TADs with a gradually increasing TAD connectivity were associated with several hematopoiesis-related functional terms. They showed strong H3K27ac, H3K4me1, and H3K4me3 signals as early as in eAECs. Specifically, H3K27ac increased significantly during the pre-HSC to LT-HSC transition, companied by a gradual reduction in H3K27me3 (Figure 2).
The most remarkable change of chromatin interaction happens at the early cell-type transition. The intra-TAD connectivity changed dramatically during eAEC to HEC transition, but altered slightly during later pre-HSC to LT-HSC transition.
HSC-related regulatory elements are pre-configured with active histone modifications at the eAEC stage. This study found that local enhancers and promoters related to hematopoiesis have already been accumulated with H3K27ac, H3K4me3 and H3K4me1 from the onset of EHT. These regulatory elements were further activated during later pre-HSC to LT-HSC transition (Figure 2).
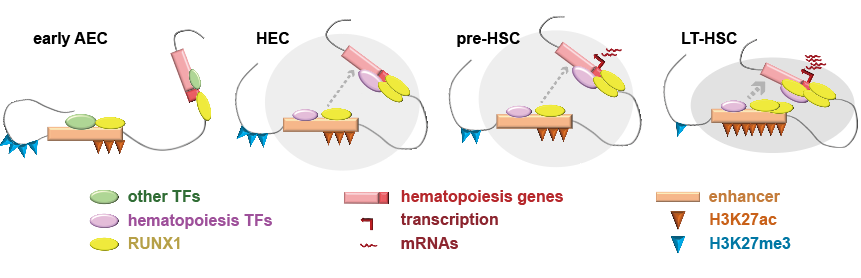
Figure 2. Multilayered epigenetic mechanisms promoting hematopoiesis
RUNX1 proteins occupy target E-P (enhancer-promoter) interactions primed with feature active histone marks before EHT. RUNX1 is as a well-known master TF essential for developmental hematopoiesis in orchestrating the EHT process5. This study examined its role in enhancer and promoter looping by performing RUNX1 itChIP-seq using as few as 500 cells for integrated analysis with Hi-C data, which was a breakthrough of RUNX1’s function in vivo . The results indicated that RUNX1-engaged E-P interactions promoted hematopoiesis early as in eAECs, which were reinforced within EHT. In addition, the study predicted TF candidate working together with RUNX1 in mediating central HSC promoter-enhancer loops (Figure 3), bringing new insights to RUNX1’s roles.
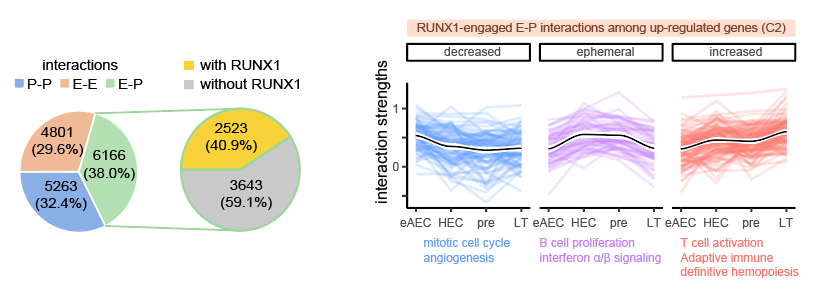
Figure 3. RUNX1-engaged E-P interactions promoting hematopoiesis
In summary, breaking the bottlenecks of cell numbers with new epigenetic detection technology (sisHi-C and itChIP), this study integrated datasets for chromatin interaction, histone modifications and RUNX1 occupancy as well as single-cell transcriptome in low-input newly-defined cell populations. The authors approached how the dynamics of multi-layered chromatin states shaped the path of HSC ontogeny.
Chen Li, Ph.D from College of Future Technology, Peking University and Guangyu Zhang, Ph.D candidate from Chinese Academy of Military Science, are co-first author of this work. Professor Aibin He (Peking University), Professor Bing Liu (Fifth Medical Center of Chinese PLA General Hospital) and Professor Yu Lan (Jinan University) direct this work. Wei Xie Lab at Tsinghua University was thanked for sharing the protocol of low-input Hi-C method. The study was supported by the Peking-Tsinghua Center for Life Sciences (by A.H.), the 1000 Youth Talents Program of China (by A.H.), and the grants from the National Key R&D Program of China (by B.L. and A.H.), the National Natural Science Foundation of China (by B.L. and A.H.). The authors thank and appreciate for the support and help from Institute of Molecular Medicine in College of Future Technology and Phoenix Platform Instrument Center in Peking University.
Aibin He Lab in Institute of Molecular Medicine, College of Future Technology, focus on single-cell epigenetic technology development and real-time single-cell imaging. Please refer to http://aibinlab.org/. Ph.D candidates and post-doctors are welcomed!
Original Link: https://www.nature.com/articles/s41467-022-28018-z
References:
1. Hou S , et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res 30, 376-392 (2020).
2. Zhou F , et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487-492 (2016).
3. Ai S , et al. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol 21, 1164-1172 (2019).
4. Du Z , et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232-235 (2017).
5. Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887-891 (2009).