The cornerstone of modern optical microscopy technology lies in eliminating scattering light and increasing optical sectioning capability within samples, such as confocal microscopy, multi-photon microscopy, light-sheet microscopy, 3D structured illumination microscopy (3DSIM), and tissue clearing methods. However, optical sectioning techniques often come with increased system costs, compromised temporal resolution, and extra phototoxicity. And biological sectioning approaches introduce complexity to the process of sampling. Moreover, even state-of-the-art optical sectioning imaging technologies still suffer from out-of-focus background caused by deep scattering when imaging thick specimens, hindering the observation of fine biological structures.
To enhance optical sectioning capabilities across diverse fluorescence imaging scenarios, a collaborative team led by Peng Xi from Peking University and Junle Qu from Shenzhen University developed an innovative approach. By integrating computer vision with fluorescence microscopy, they invented a dark channel-based optical sectioning algorithm (Dark sectioning). This research, titled "Dark-based optical sectioning assists background removal in fluorescence microscopy," was published in Nature Methods. Dark sectioning efficiently removes out-of-focus background only using a single-frame image, significantly improving signal-to-background ratio (SBR) and structural similarity (SSIM). This advancement opens new possibilities for deep tissue research, pathological diagnosis, and dynamic observation of living organisms.
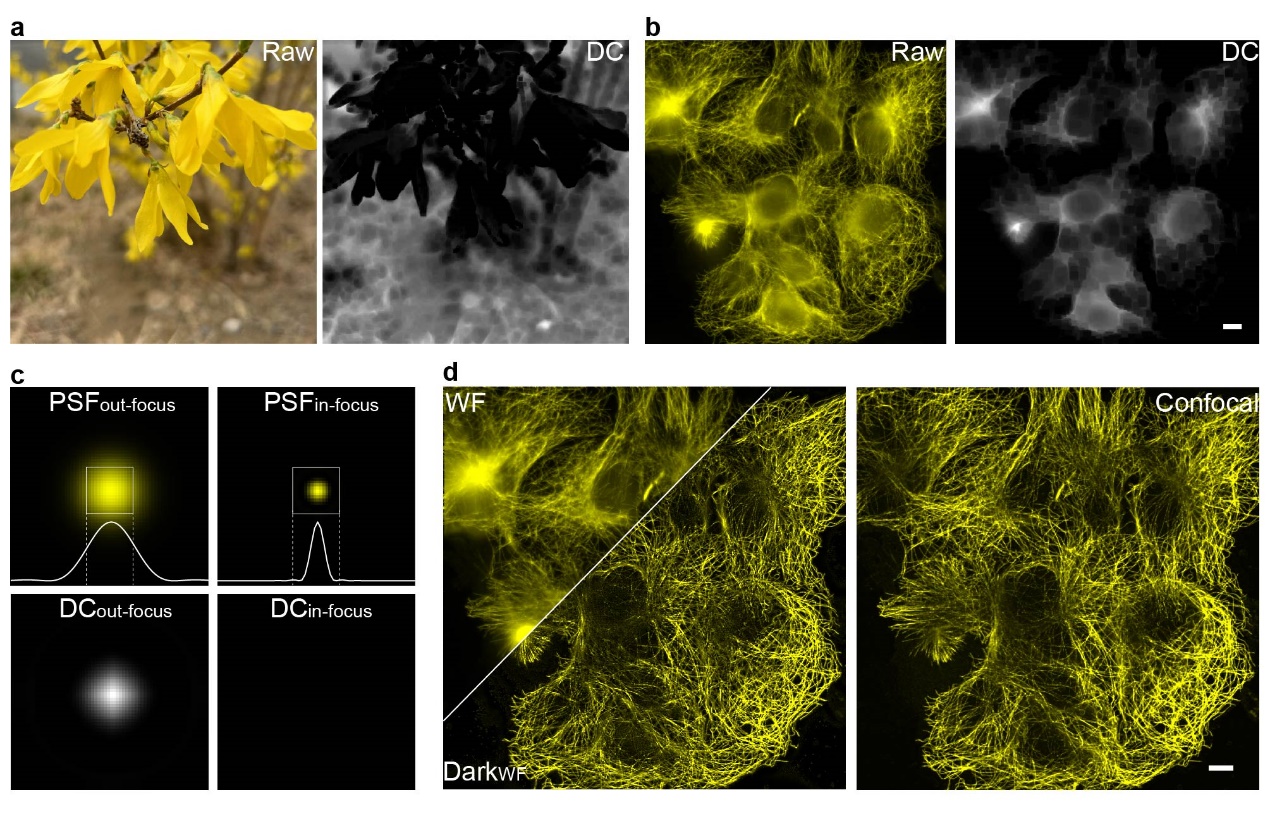
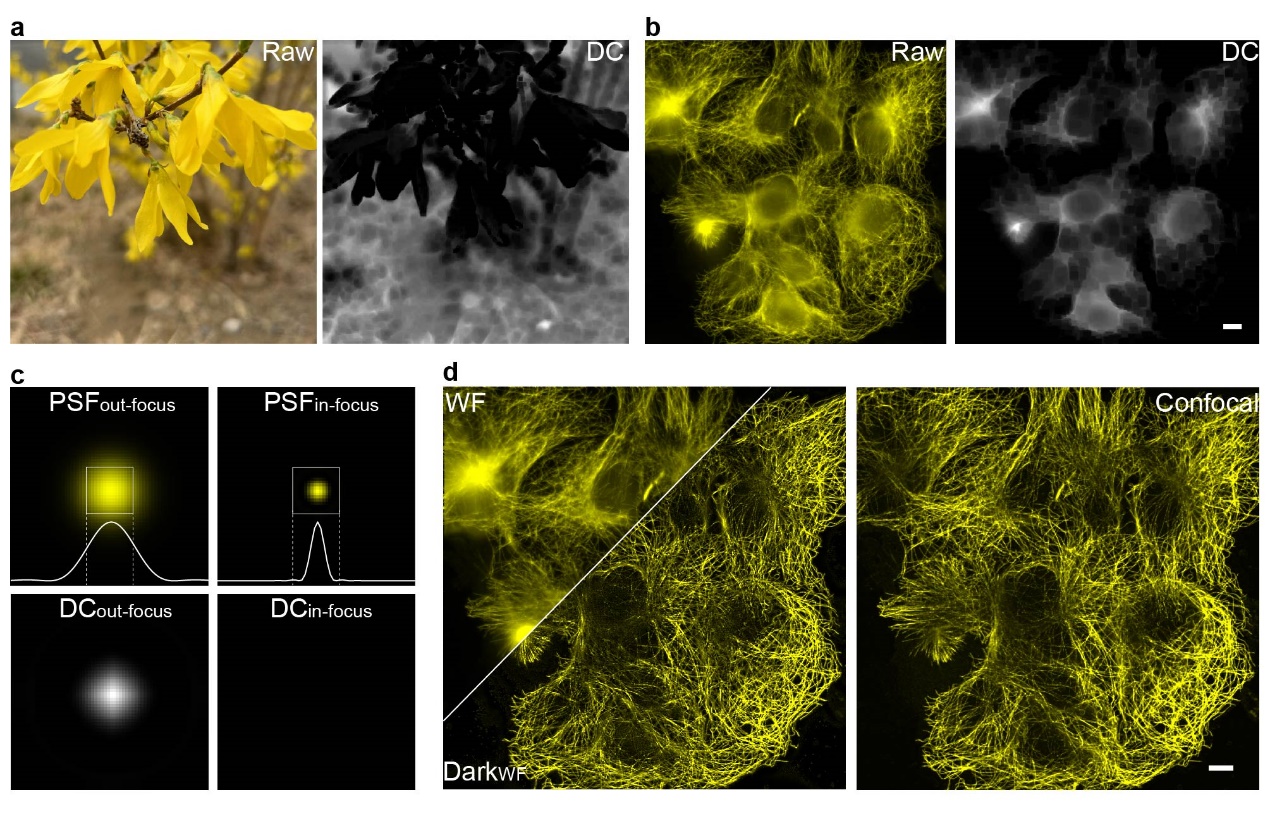
Figure 1. (a, b) Dark channel processing results for natural images and out-of-focus fluorescence images; (c) Differences in dark channel images caused by variations in point spread function (PSF) sizes; (d) Comparison of images before and after Dark sectioning. Scale bar: 4 μm.
The development of Dark sectioning was inspired by the dark channel prior used in natural image dehazing. In natural image dehazing, the dark channel prior defines an image composed of the minimum pixel values within local patches. For haze-free natural images, the dark channel appears nearly black (values close to 0), while hazy images exhibit a whitish distribution in their dark channels. This prior also distinguishes in-focus and out-of-focus regions in monochromatic images. Similarly, in fluorescence microscopy, the difference in point spread function (PSF) sizes between in-focus and out-of-focus regions allows the dark channel prior to separate foreground signals from background noise. However, critical differences exist between fluorescence images and natural haze. Haze in natural images is globally uniform, whereas out-of-focus backgrounds in fluorescence microscopy exhibit local fluctuations. Natural image pixel values represent scene information, while fluorescence pixel values directly correlate with fluorophore emission intensity. As a result, direct application of natural image dehazing to fluorescence images risks losing weak signals and incompletely removing backgrounds due to these inherent differences.
To solve the problems, the researchers developed a background removal method for fluorescence imaging called dark-channel optical sectioning (Dark sectioning). This method first separates high- and low-frequency components of the image to preserve weak signals, applying background removal only to the low-frequency part. It optimizes image patch sizes using physical point spread functions (PSFs) and simulates the initial background distribution using an extra low-pass filter to quantitatively estimate the background. Finally, iterative filtering is applied for background removal to adapt to varying background scenarios. Through cross-validation across widefield to confocal, widefield to optical-sectioning SIM, 2DSIM to 3DSIM, and one-photon to two-photon systems, the team demonstrated that Dark sectioning achieves imaging quality comparable to confocal/optical-sectioning SIM/3DSIM/two-photon microscopy using only widefield images. Moreover, it further enhances the optical sectioning capabilities of these state-of-art techniques in deep-tissue imaging.
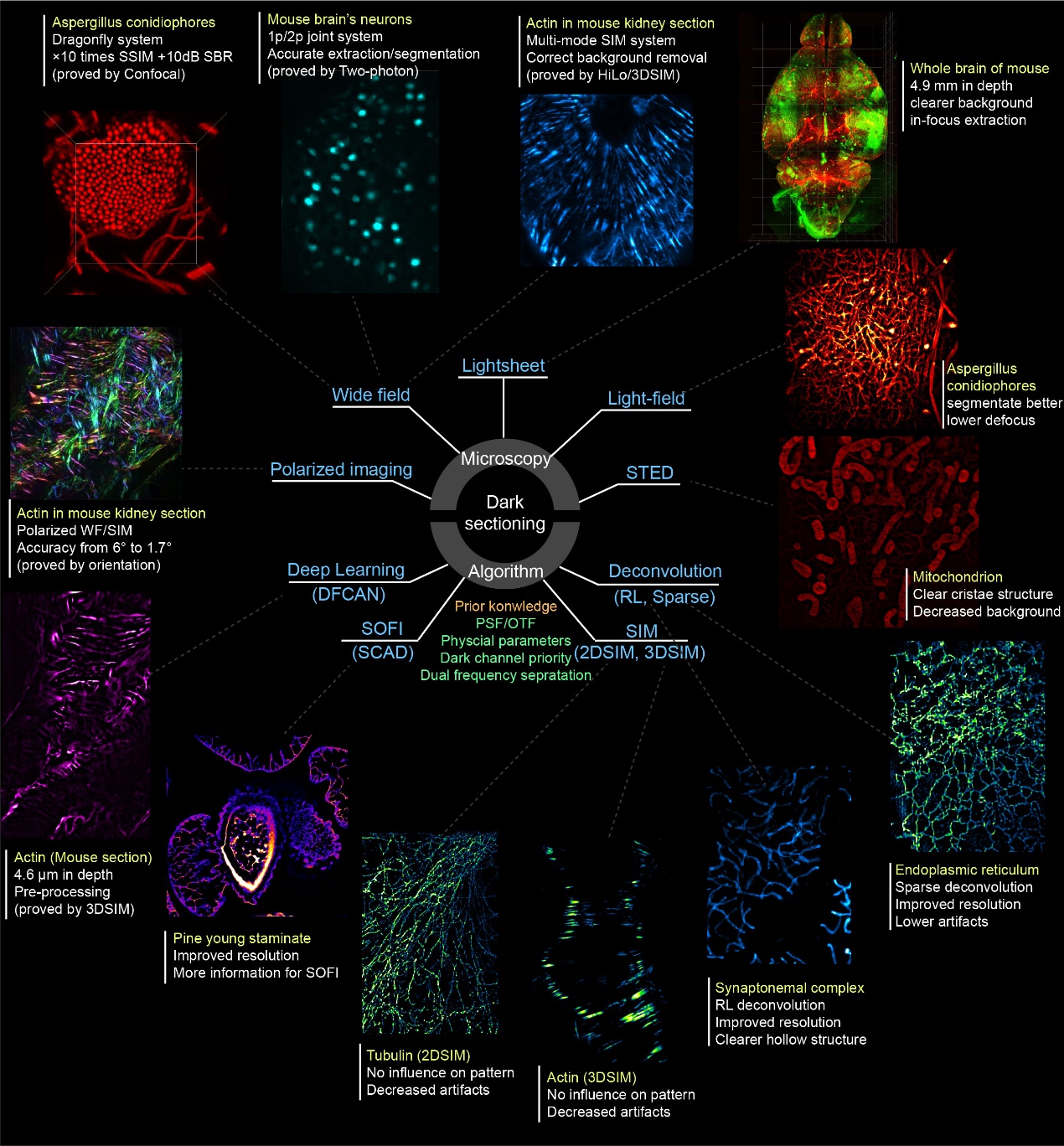
Figure 2. Dark sectioning compatibility with diverse fluorescence-based microscopy systems and reconstruction/post-processing algorithms. These include: Hardware modalities: widefield imaging, light-sheet/light-field imaging, stimulated emission depletion (STED) imaging, polarization-resolved imaging; Reconstruction/post-processing methods: deep learning-based super-resolution, super-resolution optical fluctuation imaging, 2D/3D structured illumination microscopy (SIM), and deconvolution algorithms.
Finally, the researchers systematically explored the application scenarios of Dark sectioning across diverse optical microscopy hardware modalities and reconstruction/post-processing algorithms, as illustrated in Fig. 2. They found that Dark sectioning is not only compatible with hardware modalities such as widefield, light-sheet/light-field, stimulated emission depletion (STED) imaging, and polarization imaging to enhance optical sectioning and polarization analysis capabilities; but also synergizes effectively with reconstruction/post-processing algorithms like deep learning-based super-resolution, super-resolution optical fluctuation imaging (SOFI/SACD), 2D/3DSIM), and deconvolution. The technique significantly reduces artifacts in 2D/3D SIM reconstructions while improving the resolution of SOFI and deconvolution.
By innovatively combining computer vision with fluorescence microscopy, Dark sectioning achieves enhanced sectioning performance across various imaging modalities without additional hardware costs. In the future, it could be integrated into real-time out-of-focus removal workflows for microscopes and serve as a universal preprocessing tool for reconstruction and deep learning. Notably, due to its simplicity and computational efficiency, Dark sectioning operates at high speeds with strong generalizability. To improve accessibility, the team has released multi-platform tools including MATLAB scripts, Java-based Fiji plugins, and standalone executables for public testing: https://github.com/Cao-ruijie/Dark-sectioning.
Following its publication, this work has been received positive feedbacks from renowned scholars world-widely. Prof. Emmanuel Soubies (CNRS researcher and founder of FlexSIM) praised the work and integrated Dark sectioning into FlexSIM, commenting on Bluesky: "Nice work! We integrated this dark-sectioning method into our FlexSIM pipeline as an additional option for handling out-of-focus signal. It works remarkably well!" Dr. Rita Strack, a senior editor of Nature Methodshas also highly commented this work: “A number of useful methods have been established for denoising fluorescence microscopy images. However, removing out-of-focus background signal is still challenging. I was impressed by how these authors ingeniously borrowed ideas from dehazing of natural images and applied them to fluorescence microscopy to remove background signal and dramatically improve image quality.” Prof. Biagio Mandracchia from University of Valladolid, Spain has also given his opinion on this work: “Overall, dark sectioning is a novel, widely applicable solution to a persistent problem in fluorescence microscopy. Dark sectioning has the potential to substantially advance biological imaging and research by allowing microscopists to match confocal-like imaging with much more accessible and faster wide-field microscopes.”
In this study, Prof. Peng Xi (Peking University's Future Technology Institute) and Prof. Junle Qu (Shenzhen University) served as corresponding authors. Ruijie Cao (PhD candidate, Peking University) and Yaning Li (graduated with PhD from Peking University) contributed equally as co-first authors. The work also received critical support from research groups led by Prof. Peng Fei (Huazhong University of Science and Technology), Prof. Jiamin Wu (Tsinghua University), Prof. Yiming Li (Southern University of Science and Technology), Prof. Changhui Li (Peking University), and Prof. Yanye Lu (Peking University). This work was funded by the National Key R&D Program of China and the National Natural Science Foundation of China.
Link to the paper:https://doi.org/10.1038/s41592-025-02667-6
Nature Methods Research Briefing: https://doi.org/10.1038/s41592-025-02668-5